My boss sent me an email (on my day off!) asking me just how much of R is written in the R language. This is very simple if you use R and a Unix-like system. It also gives me a good excuse to defend the title of this blog. It’s librestats, not projecteulerstats, afterall.
So I grabbed the R-2.13.1 source package from the cran and wrote up a little script that would look at all .R, .c, and .f files in the archive, record the language (R, C, or Fortran), number of lines of code, and the file the code came from; then it’s just a matter of dumping all that to a csv (converted to .xls (in LibreOffice) because Wordpress hates freedom).
We’ll talk in a minute about just how you would generate that csv–but first let’s address the original question.
By a respectable majority, most of the source code files of core R are written in R:
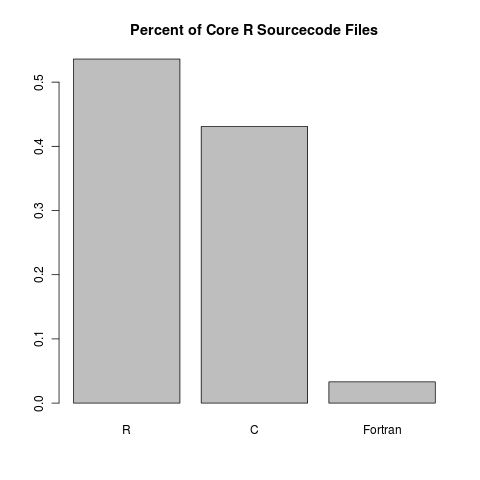
At first glance, it seems like Fortran doesn’t give much of a contribution. However, when we look at the proportion of lines of code, we see something more reasonable:
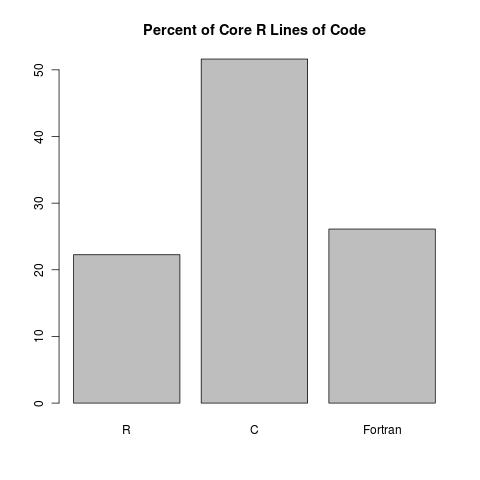
So there you have it. Roughly 22% of R is written in R. I know some people want R to be written in R for some crazy reason; but really, if anything, that 22% is too high. Trust me, you really want C and Fortran to be doing all the heavy lifting so that things stay nice and peppy.
Besides, this is a fairly irrelevant issue, in my opinion. What matters is that people outside of Core R are writing in R. Look at the extra packages repo and you’ll see a very different story from the above graphic. That’s something SAS certainly can’t say, since people who want to do anything other than call some cookie-cutter SAS proc have to use IML or that ridiculous SAS macro language–each of which is somehow even more of a hilarious mess than base SAS.
Ok, so how do we get that data? I actually have a much better script than the one I’m about to describe. The new one automatically grabs every source package from the cran that you don’t already have and starts digging in on them, dumping everything out into one big csv so you can watch trending. It’s interesting to see the transition from R being almost entirely (92%) in C to seeing it slowly drop down to ~52%. But that’s a different post for a different day because I have a few kinks to work out with that script before I would feel comfortable releasing it.
So here’s how this system works. It’s basically the dumbest possible solution; I’m pretty good at those, if I may say so myself. Basically the shell script hops into across the R-version/src/ folder and gets a line count of each .R, .c, and .f file. That’s it; here it is:
#!/bin/sh
outdir="/path/to/where/you/want/the/csv/dumped"
rdir="/path/to/R/source/root/directory/to/be/examined" #eg, ~/R-2.13.1/
cd $rdir/src
for rfile in `find -name *.R`
do
loc=`wc -l $rfile | sed -e 's/ ./,/' -e 's/\/[^/]*\//\//g' -e 's/\/[^/]*\//\//g' -e 's/\/[^/]*\///g' -e 's/\///'`
echo "R,$loc" >> $outdir/r_source_loc.csv
done
for cfile in `find -name *.c`
do
loc=`wc -l $cfile | sed -e 's/ ./,/' -e 's/\/[^/]*\//\//g' -e 's/\/[^/]*\//\//g' -e 's/\/[^/]*\///g' -e 's/\///'`
echo "C,$loc" >> $outdir/r_source_loc.csv
done
for ffile in `find -name *.f`
do
loc=`wc -l $ffile | sed -e 's/ ./,/' -e 's/\/[^/]*\//\//g' -e 's/\/[^/]*\//\//g' -e 's/\/[^/]*\///g' -e 's/\///'`
echo "Fortran,$loc" >> $outdir/r_source_loc.csv
done
Then the R script just does exactly what you’d think, given the data (take a look at the “csv” for examples).
r.loc <- read.csv("r_source_loc.csv",header=FALSE)
a <-r.loc[which(r.loc[1] == "R"),][2]
b <-r.loc[which(r.loc[1] == "C"),][2]
c <-r.loc[which(r.loc[1] == "Fortran"),][2]
files.total <- length(a[,1])+length(b[,1])+length(c[,1])
loc.total <- sum(a)+sum(b)+sum(c)
cat(sprintf("\nNumber .R source files:\t\t %d\nNumber .c source files:\t\t %d\nNumber .f source files:\t\t %d\n",length(a[,1]),length(b[,1]),length(c[,1])))
cat(sprintf("-------------------------------------"))
cat(sprintf("\nTotal source files examined:\t %d\n\n",length(a[,1])+length(b[,1])+length(c[,1])))
cat(sprintf("\nLines of R code:\t %d\nLines of C code:\t %d\nLines of Fortran code:\t %d\n",sum(a),sum(b),sum(c)))
cat(sprintf("-------------------------------"))
cat(sprintf("\nTotal lines of code:\t %d\n\n",loc.total))
cat(sprintf("\nAmong all lines of code being either R, C, or Fortran:\n"))
cat(sprintf("%% code in R:\t\t %f\n%% code in C:\t\t %f\n%% code in Fortran:\t %f\n",100*sum(a)/loc.total,100*sum(b)/loc.total,100*sum(c)/loc.total))
png("pct_r_source_files.png")
barplot(c(length(a[,1])/files.total,length(b[,1])/files.total,length(c[,1])/files.total),main="Percent of Core R Sourcecode Files",names.arg=c("R","C","Fortran"))
dev.off()
png("pct_r_code.png")
barplot(c(100*sum(a)/loc.total,100*sum(b)/loc.total,100*sum(c)/loc.total),main="Percent of Core R Lines of Code",names.arg=c("R","C","Fortran"))
dev.off()
From the R script, we can get precise figures, which I prefer to pictures any day. But I seem to be an outlier in this regard…
Number .R source files: 729
Number .c source files: 586
Number .f source files: 45
-------------------------------------
Total source files examined: 1360
Lines of R code: 149520
Lines of C code: 346778
Lines of Fortran code: 175409
-------------------------------
Total lines of code: 671707
Among all lines of code being either R, C, or Fortran:
% code in R: 22.259705
% code in C: 51.626379
% code in Fortran: 26.113916